Image Analysis
Image data can be analyzed by our user in the computer room U04. Currently we are in the process of upgrading to new analysis computers, so that at the moment only two workstations are available for our users. Since we do not have a position for an image analysis specialist, we can only offer limited help with data analysis. The following programs are available:
- NIS-Elements AR (from Nikon)
- Volocity (from PerkinElmer)
- Huygens (from SVI)
- ImageJ/Fiji
NIS-Elements in the "Advanced Research" (AR)-version from Nikon is the central program for image acquisition and analysis in the NIC. It handles multi-dimensional imaging flawlessly, with support for capture, display, peripheral device control, data management and analysis of up to six dimensions (X,Y,Z, Lambda (wavelength), T, multipoint). It also offers sophisticated image processing features, such as an extremely powerful deconvolution module, exclusive one-click database capability, and Extended Depth of Focus function. All users of the NIC may use NIS-Elements as analysis software on several workstations in the center.
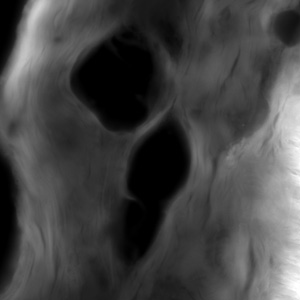
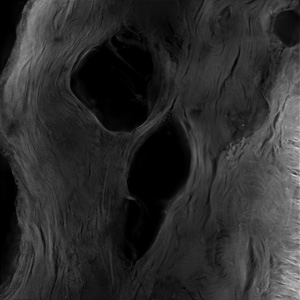
Volocity from PerkinElmer is the acquisition software used for our spinning disk microscope (VoX-Ti). Apart from image acquisition we're also offering the software in a analysis license.
Huygens from Scientific Volume Imaging (SVI) is a powerful deconvolution software that is controlled by a web interface (the Huygens Remote Manager) which means that you may up- and download your data and control Huygens from outside the NIC. It runs on a server with 2 10-core Xeon CPUs and 1 TByte of RAM. It can be used in a batch mode, deconvolving many big data files sequentially without user interaction.
Huygens usage is free for all microscope users in the NIC after a short introduction to the program on request (free of charge). Others may get access to Huygens for an annual fee of 80€ (please contact me). The Huygens Remote Manager may be reached via http://bambam.bioquant.uni-heidelberg.de/hrm/.
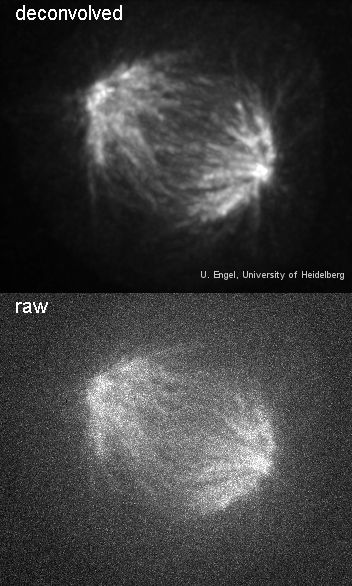
Spindle before and after deconvolution
Users who prefer to analyze their data acquired in the NIC in their own lab may use the free software Fiji, that is able to open data from NIS-Elements or Volocity (this requires a plugin called "Bio-Formats" that is included with Fiji).
A tutorial describing Fiji, written by Peter Bankhead, former image analysis specialist of the NIC, can be downloaded here. In case of questions you may contact Pete by e-mail.



